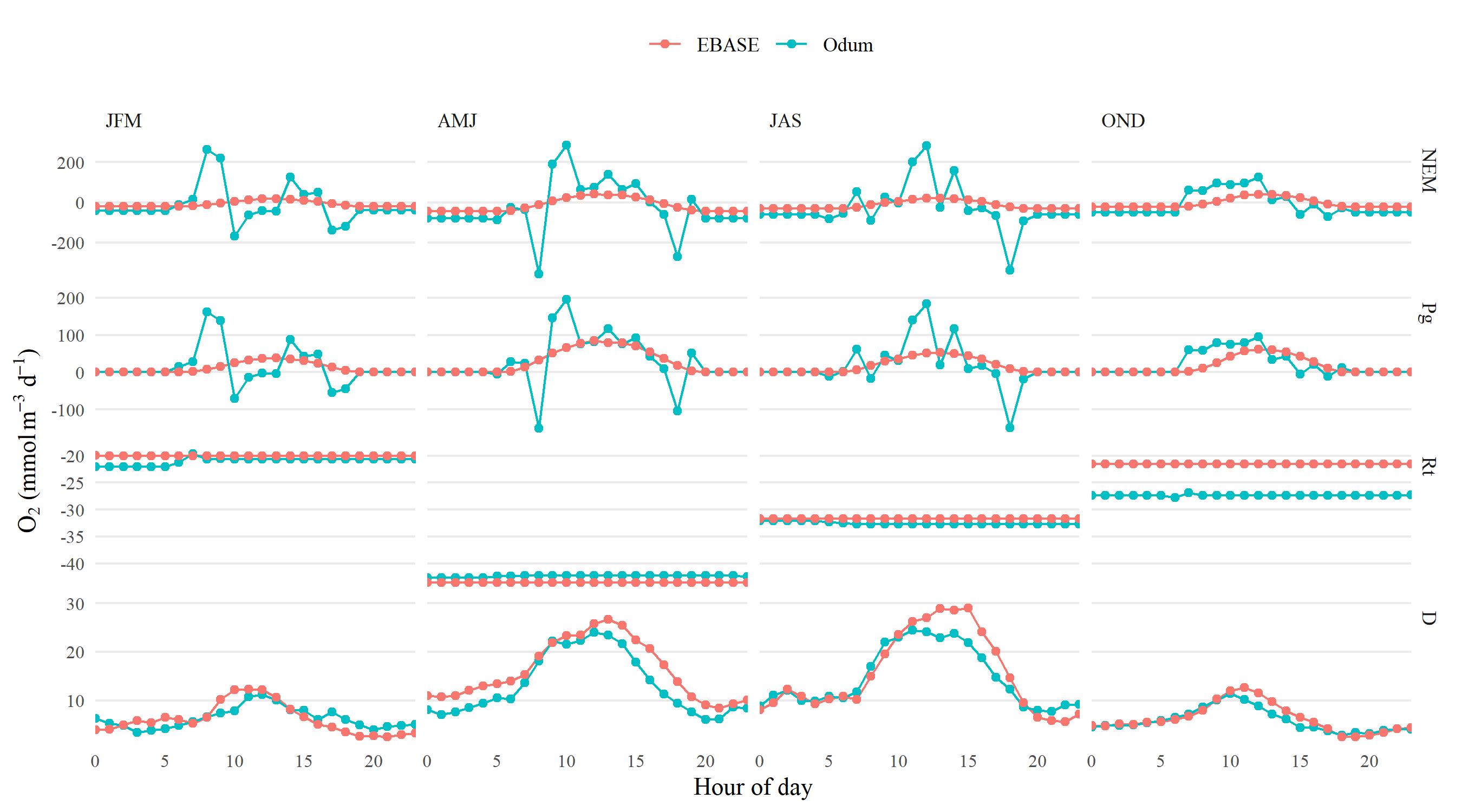
EBASE vs Odum metabolism estimates
Summary
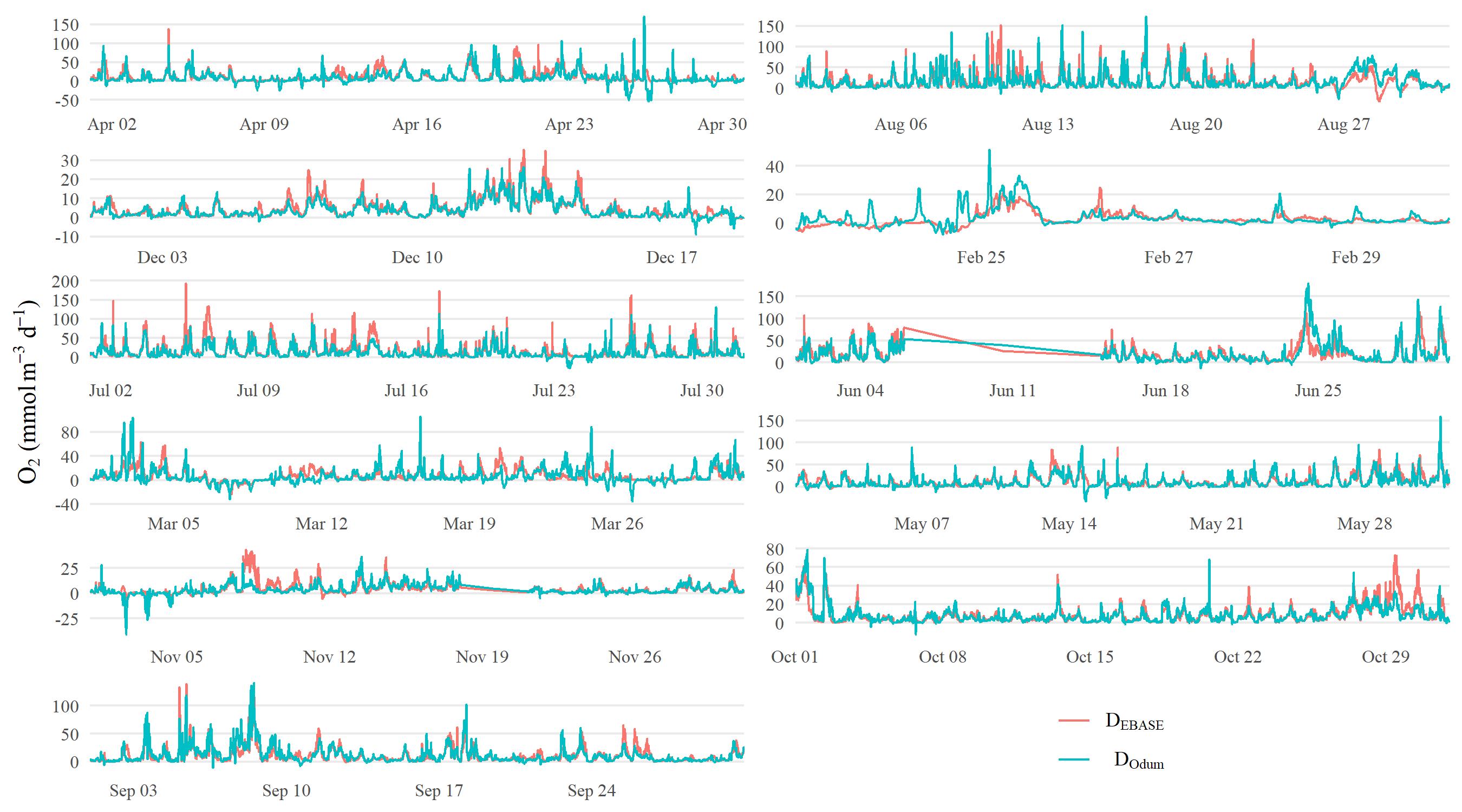
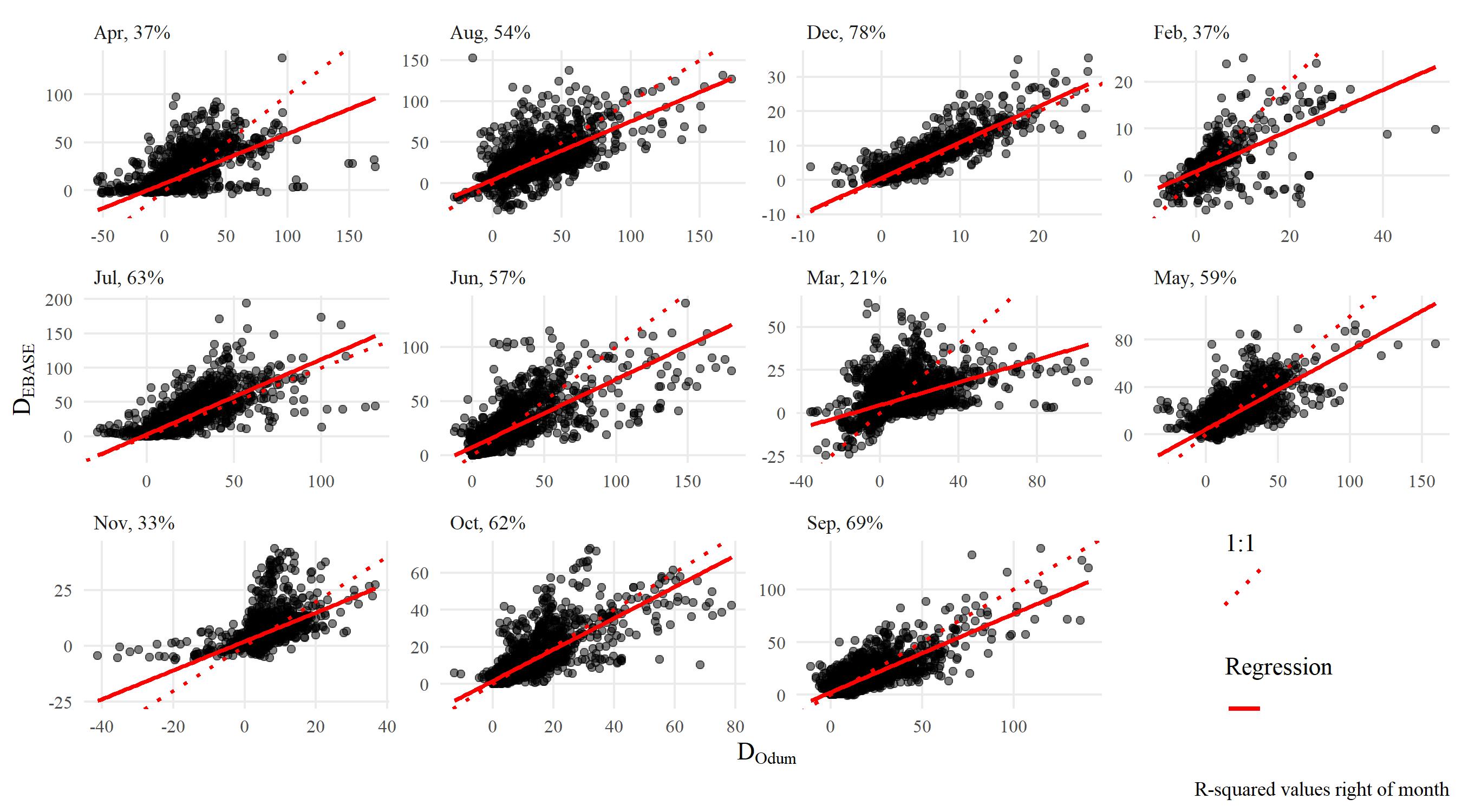
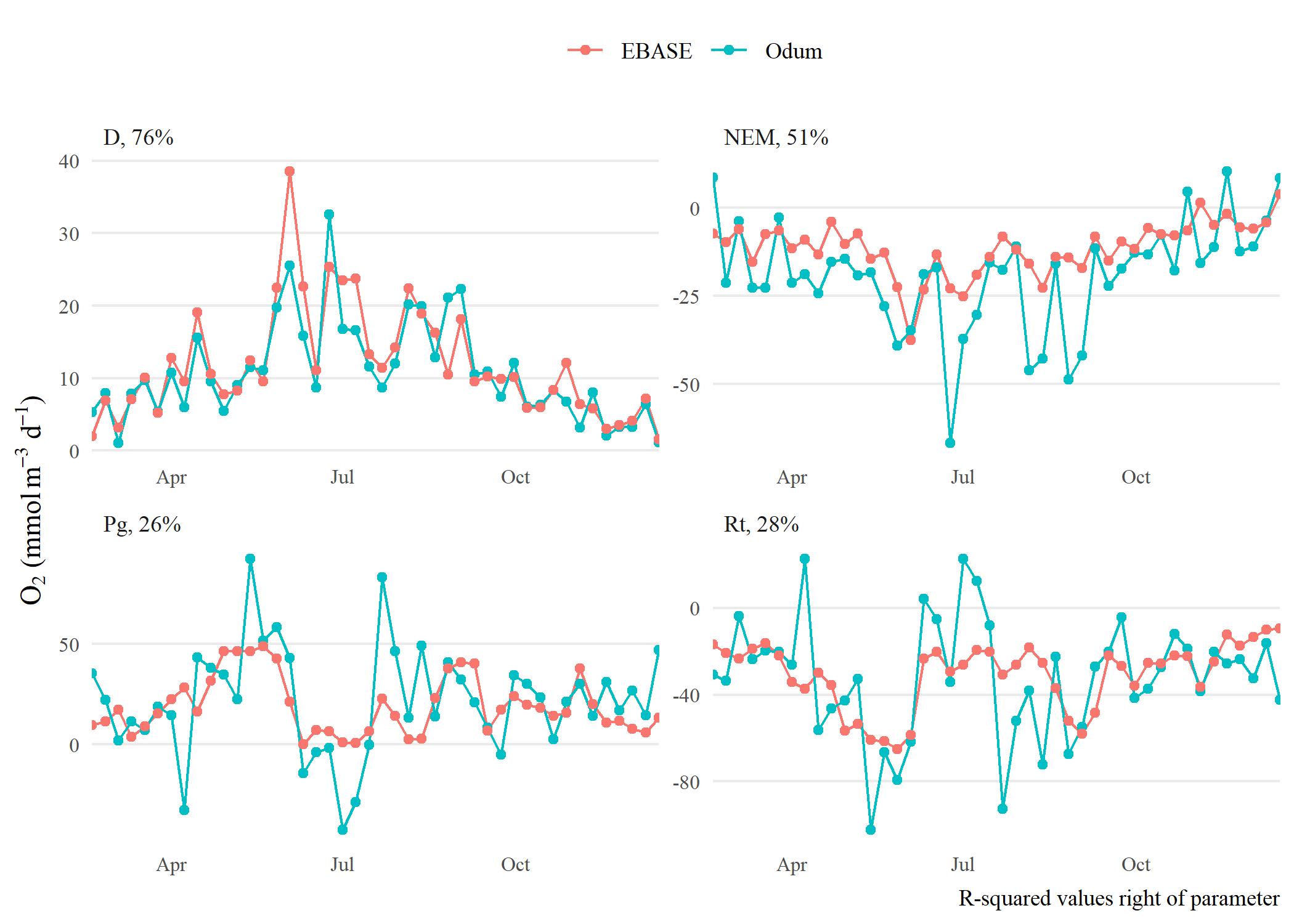
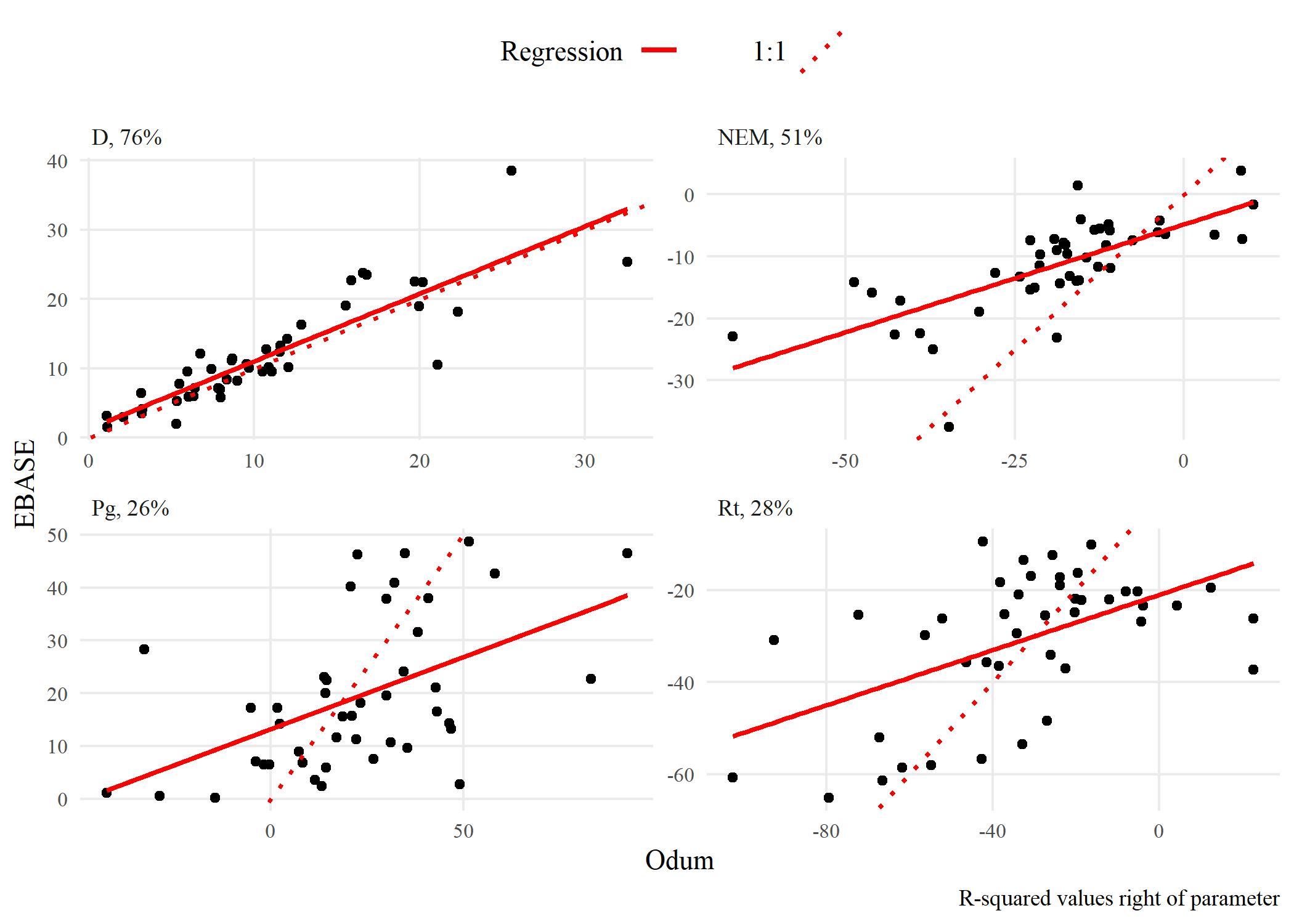
The following shows results comparing metabolic estimates obtained from the EBASE and Odum models. The EBASE model was run using the EBASE R packages and the Odum model was run using the ecometab function from the WtRegDO R package.
All models were run using the observed dissolved oxygen time series for data from 2012 at Apalachicola NERR. The EBASE model was set to optimize the parameter equation over a seven day period. Both models used the Wanninkhof equation to estimate the gas exchange. The source code used to create the models and all figures below is in the link above.